| |
| |
Группа моделирования белковых структур |
|
| |
Заведующий – А.В. Ефимов
1. Several novel super-secondary structures or structural motifs having unique overall folds have been revealed and described. In globular proteins, the polypeptide chain folds over upon itself many times and the elements of secondary structure that are close together in the sequence have a strong tendency to be in close contact in three-dimensional space. Very often, α-helices and/or β-strands that are adjacent along the chain form a few well-defined types of structural motifs. While many different structural motifs have been observed to recur within globular proteins, only some of the motifs exhibit unique handedness and a unique overall fold. Structural motifs of a given type found in unrelated proteins may have the same overall fold despite their α-helices and/or β-strands being of different length, their connection regions differing in length and conformation and their sequences lacking homology. The structural motifs with unique overall folds can act as nuclei or «ready-made» building bloks in protein folding. Alternatively, the motifs may be regarded as the starting structures for protein modeling and prediction studies and can be used to search for all possible protein folds. Some examples of such structural motifs are given in Fig.1.
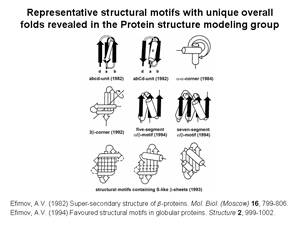
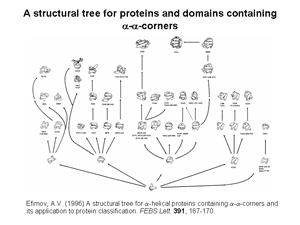
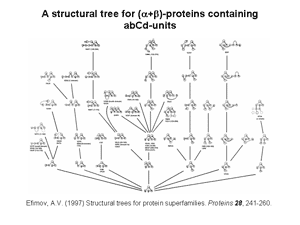
2. Nine structural trees for the largest protein superfamilies have been constructed and analyzed. The structural motifs having unique overall folds and a unique handedness are taken as the starting structures in modeling or as the root structures of the trees. The larger protein structures of each superfamily are obtained by stepwise addition of α-helices and/or β-strands to the root structural motif taking into account a restricted set of rules inferred from known principles of the protein structure. Among these rules, prohibition of crossing connections, attention to handedness and compactness, and a requirement for α-helices to be packed in α-helical layers and β-strands in β-layers are the most important. A scheme that includes the root structural motif, all the intermediate and final structures connected by lines showing possible folding pathways is represented as a structural tree. The structural trees are a good tool for searching of all possible protein folds and folding pathways as well as for structure comparision and protein classification. Figs 2 and 3 show two representative structural trees for α-helical and (α+β)-proteins.
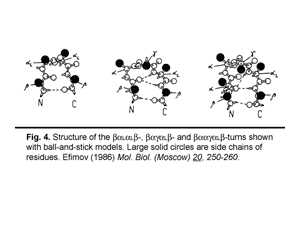
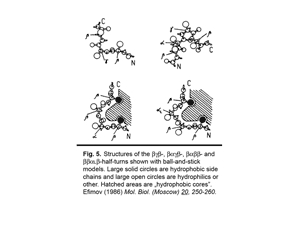
3. Classification of protein irregular regions and shorthand nomenclature for description of the conformation of irregular regions have been developed. There are four types of loop regions if they are classified according to the secondary-structural elements they connect: αα, αβ, βα and ββ. Irregular regions, especially short ones, can be grouped into two classes according to their «function»: turns providing for the polypeptide chain reverse turns by 180° and half-turnes to change the polypeptide chain trace by approximately 90°. A number of novel small standard structures in irregular regions of proteins, including turns, half-turns, N- and C-caps of α-helices etc. have been revealed as a result of stereochemical analysis. It was shown that long loops in proteins can be represented as combinations of the small standard structures. Some representative turns are shown in Fig.4 and half-turnes in Fig.5. To describe the conformation of turns and half-turns a shorthand nomenclature is used. It has been derived from the observed φ, ψ distributions in the Ramachandran plot where six regions α, α(L), β, γ, δ and ε containing the great majority of the experimental points (φ, ψ-pairs) can be distinguished.
|
|
| |
|
|
| |
Группа химии пептидов |
|
| |
Заведующий – Ю.В. Митин
В природе существуют антимикробные пептиды, которые эффективно убивают бактерии. Антимикробные пептиды взаимодействуют с мембранами бактерий в отличие от антибиотиков, которые действуют на отдельные стадии метаболизма. В аминокислотной последовательности ряда антимикробных пептидов встречается тетрапептидные фрагменты, KFGK (магаинин), RLLR (буфорин), состоящие из двух положительно заряженных аминокислот, между которыми находится одна или две гидрофобные аминокислоты. Такие тетрапептидные фрагменты являются основными структурными единицами природных антимикробных пептидов, при этом в пептидной цепи содержится несколько фрагментов такого типа. Мы синтезировали тетрапептиды RLAR, и KLAK, которые сами по себе не активны, но на их основе можно получить как линейные [1], так и разветвленные пептиды [2], обладающие антимикробной активностью. Были синтезированы также разветвленные пептиды E[E(RLAR)2]2, E[E(KLAR)2]2, активность которых была близка к активности таких природных антимикробных пептидов, как магаинин и темпорин: 10-15μМ (по отношению к E.coli.).
|
|
| |
|
|
| |
Персональная группа В.Д. Васильева |
|
| |
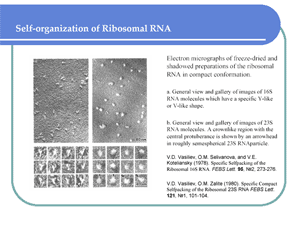
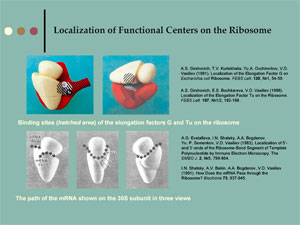
|
|
| |
|
|
| |
Персональная группа Ю.Н. Чиргадзе |
|
| |
Crystal structure of elongation factor G from Thermus thermophilus has been determined at 2.85 Å. Protein molecule has a complex structure, it consists of five domains, and the first domain is carrying GTPase activity. This work has been carried out in collaboration with the group of Dr. Garber, Inst.Prot.Res. and Lab. of Prof. A. Liljas, Univ. of Lund, Sweden.
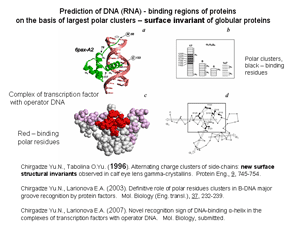
Functional important and evolutionary conservative alternating charge clusters of amino acid side groups have been discovered on the surface of bovine eye lens gammaD-crystallin. This is one of the first surface structural invariant in addition to variety invariants known for the internal structure of protein molecule. Similar charge clusters have been found on the surface of 86% of globular proteins with known spatial structure.
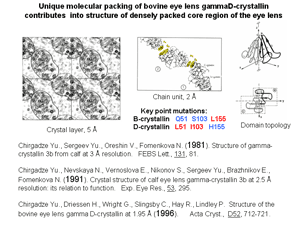
It was shown that in complexes of protein with DNA and RNA the contact regions consist of the side groups of polar residues which concentrate in one largest cluster on the protein surface. This suggests that large clusters of polar residues play a key role in formation of both DNA- and RNA-binding sites of proteins. In accordance with this for 65 complexes of transcription factors with fragments of operator B-DNA the polar residues of binding helix display important distinctive sign of recognition: in 96% of binding protein factors binding polar residues are disposed inside one of two largest clusters on the protein surface.
|
|
| |
|
|
|
|
