Заведующий – А.В. Финкельштейн
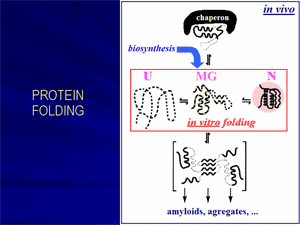
Originally inspired mainly by a scientific curiosity, protein folding problem has now become of primary medical importance, given that incorrect folding of proteins is now known to be responsible for many devastating diseases.
«Molten globule»: the main protein folding intermediate.
How does a protein chain find its unique structure among zillions of alternatives? This fundamental problem aroused when the very first protein structures have been solved and it was shown that these rather complicated structures can, nevertheless, fold spontaneously not only in vivo, in vitro as well. In an attempt to solve this problem, Oleg Ptitsyn suggested (in 1973) a «sequential mechanism» of protein folding.
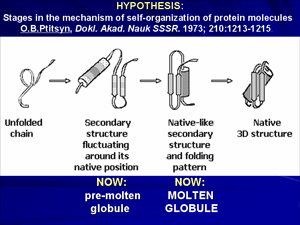
A keystone of this concept was now conventional, but that time purely hypothetical «molten globule», and its precursor, a «pre-molten globule». Molten globule, the third physical state of the protein molecule, and then the pre-molten globule has been experimentally discovered and characterized in details in our laboratory in 1980-s. Molten globule is similar to native protein in some respects, and to unfolded state in the others. This discovery allowed understanding of physical nature of protein de- and renaturation.
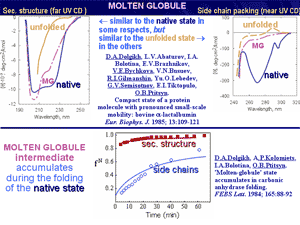
Further, it has been proposed and experimentally demonstrated that molten globules can be involved in some physiological processes and maybe even in some genetic diseases. It was shown that the molten globule state can be formed at mild denaturing conditions in the cell, e.g., at the protein-membrane interactions.
Protein folding rate theory: solution of the famous «Levinthal paradox».
The Levinthal paradox stressed that the most stable chain fold cannot be found among a plethora of alternatives by an exhaustive enumeration of possibilities (which takes more than the Universe lifetime). Therefore, for decades it was thought that not only the native fold, but a specific pathway to it also must be encoded by the amino acid sequence; consequently, it was thought that design of artificial proteins must include design of their folding pathways, which makes this design very cumbersome. However, a theoretical analysis of folding and unfolding pathways have shown that the most stable chain fold is automatically a focus of rapid folding pathways, when the chain's amino acid sequence provides a large «energy gap» between the native fold and its competitors.
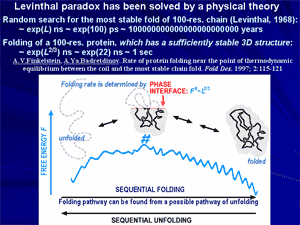
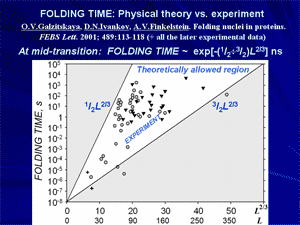
Then the chain folds via the first order phase transition (already observed by Privalov), and a nucleation process typical of these transitions brings this chain to its native fold rapidly and unambiguously. As a result, a general theory of protein folding has been developed, which paved a way to modern theories and algorithms searching the protein folding nuclei and calculating protein folding rates.
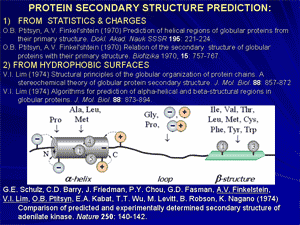
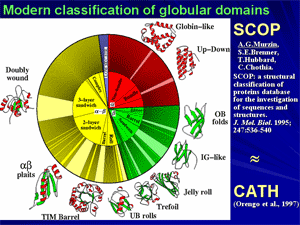
Discovery of physical reasons for existence of a narrow set of typical protein folds: a basis for rational classification of protein structures.
At the end of 1970-th it has been shown that proteins, or rather their domains, often have similar folds, even if they are quite different from functional or phylogenetic points of view. This suggests that a reason of such similarity is some physical limitation of the set of possible folds rather than functional convergence of evolutionary divergence. Structural analysis of typical folds has shown that they look rather simple and regular, because their cores are built of crystal-like stacks of secondary structures and do not have «defects» like hydrogen bonds lost within the protein body and overcrossed or too bent irregular loops.
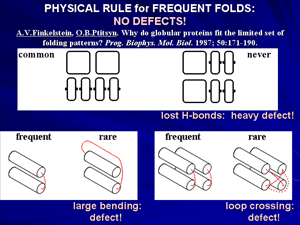
Theoretical analysis led to conclusion that typical protein folds form a narrow set of «defect-free» structures, which have a construction that facilitates stability of these folds for a vast majority of amino acid sequences.
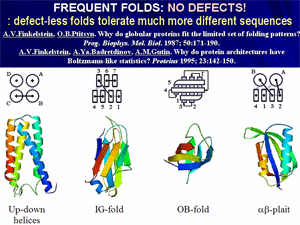
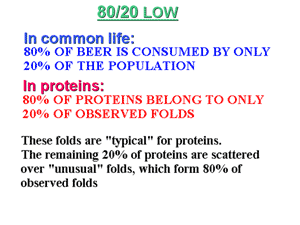
Further, this discovery led to a modern physical (or rational, as it is called now) classification of protein structures and led to a generalization, colloquially known as «80/20 law for proteins».
|
